library(TENxPBMCData)
tenx_pbmc4k <- suppressMessages(TENxPBMCData(dataset = "pbmc4k"))
mean(counts(tenx_pbmc4k) == 0)[1] 0.9608314Thomas Sandmann
December 23, 2024
I am always looking for ways to communicate genomics analyses to collaborators in a way that allows them to drill down into the data themselves. Stand-alone HTML reports can be opened in any web browser and can support rich visualizations, making them my an indespensible tool in my tool kit.
Yet, some of the data I am working with, e.g. single-cell RNA-seq data, is pretty large, and the size of my HTML reports - which include the data as text - can quickly balloon. Luckily, there are ways to compress numerical data without loosing information.
Here, I am exploring two approaches: run-length encoding (RLE) and Lempel-Ziv-Welch (LZW) compression1.
Single-cell RNA-seq data are notoriously sparse, e.g. the majority of counts across genes & cells is zero. For example, 96% of all counts in the PBMC 4k single-cell experiment shared by 10X Genomics (and available via the TENxPBMCData Bioconductor package) are zero:
[1] 0.9608314Most of the remaining counts are integers smaller than ten, with fewer than 0.2% of values exceeding that range:
This leads to very long runs of identical integers (most often zero), which can efficiently be represented through run-length encoding
For example, the expression of the transcription facto MYC gene is highly skewed towards zero counts:
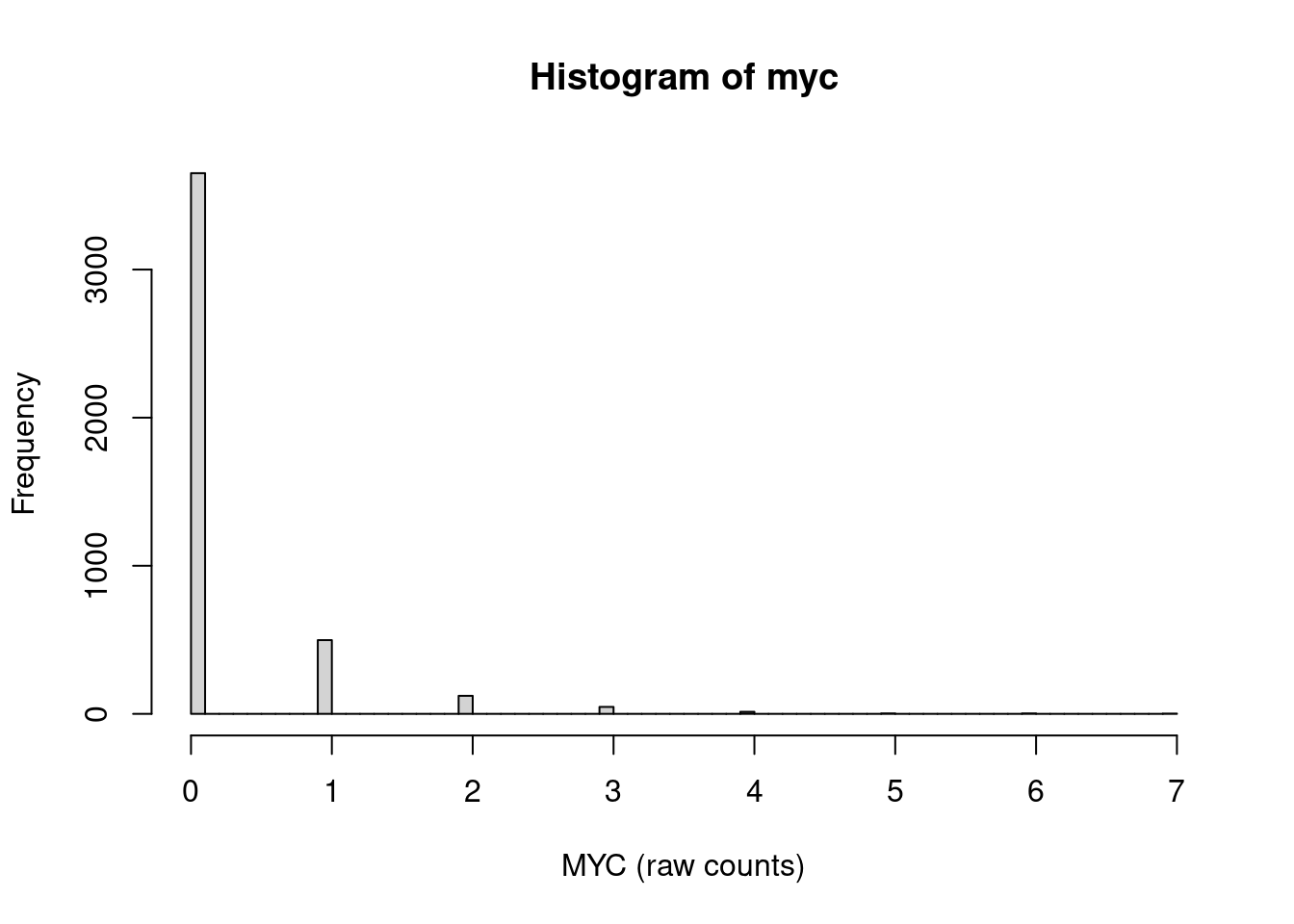
and there are long sequences of zeros and some shorter sequences of ones in the count vector. (Here, I am only showing counts for the the first 100 of the 4340 cells.)
[1] 0 0 0 1 3 0 0 0 1 0 0 0 0 1 1 1 0 0 0 1 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0
[38] 0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 2 0 1 0 0 0
[75] 0 0 0 1 0 0 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 1 0 1 0R estimates that the entire vector of 4340 counts occupies 17 KiB of memory.
This information can be communicated more concisely by noting each integer (e.g. zero) and the number of times it is repeated. The stats::rle function performs this conversion:
Run Length Encoding
lengths: int [1:1195] 3 1 1 3 1 4 3 3 2 14 ...
values : int [1:1195] 0 1 3 0 1 0 1 0 1 0 ...and returns two components:
This allows us to compress the original 4340 integers into 2390 integers (1195 length & value pairs), without losing any information.
By combining information about values and run-lengths, we can reconstruct the original vector, either manually:
0 x 3: 0,0,0,
1 x 1: 1,
3 x 1: 3,
0 x 3: 0,0,0,
1 x 1: 1,
0 x 4: 0,0,0,0,
1 x 3: 1,1,1,
...
=> 0,0,0,1,3,0,0,0,1,0,0,0,0,1,1,1,...or via inverse.rle:
To store run-length encoded values e.g. in a text (or HTML) file, we can also represent them as characters, similar to our manual reconstruction above, e.g. separating values and lengths with an x:
[1] "0x3;1x1;3x1;0x3;1x1;0x4;1x3;0x3;1x2;0x14"This representation occupies only 4.9 KiB of memory - but half of the character string is required to encode the x and ; separators. We can do better!
To avoid including any separators, we can represent one of the numbers, e.g. the values, as a single character instead. For the MYC gene, the largest observed integer count is 7. So let’s represent all 8 values (including 0) with a letter of the alphabet instead:
0 1 2 3 4 5 6 7
"a" "b" "c" "d" "e" "f" "g" "h" For example, the letter a represents 0, b represents 1 and so on.
Now we can omit the x and ; separators, as each run is unambiguously represented by one character and an integer 2:
myc_rle_character <- mapply(paste0,
1 dictionary[as.character(run_lengths$values)],
run_lengths$length) |>
paste(collapse = "")
substr(myc_rle_character, 1, 21)[1] "a3b1d1a3b1a4b3a3b2a14"In this representation, the 4340 counts value for the MYC gene now occupy only 2.6 KiB of memory instead of the original 17 KiB. We can also write the data to a text (or HTML) file in this format, e.g. as a single string.
In this example, we only needed 8 letters to encode all observed unique counts for the MYC gene. But for other genes, especially those expressed at higher levels, we might observe many more possible values - even more than the 26 lower case letters of the English alphabet.
For example, in this dataset we observe 59 distinct count values for the highly abundant GAPDH gene:
gapdh
0 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19
485 683 708 572 430 302 216 146 118 122 85 56 58 44 40 22 24 25 16 13
20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39
18 15 11 10 13 4 6 4 10 4 8 4 4 7 6 1 5 7 2 2
40 41 42 44 46 48 49 50 52 53 54 56 57 61 70 75 85 92 181
5 4 1 4 1 2 1 2 2 1 2 1 1 2 1 1 1 1 1 Luckily, we are not restricted to the English alphabet. For example, we can map to any character in the Unicode Transformation Format – 8-bit (UTF-8) character encoding instead.
To encode e.g. all integers from 1 to 256 (more than enough for GAPDH) - but avoiding those that represent a space, quote, etc or integers - we can use UTF-8 codes 64 to 319:
0 1 2 3 4 5 6 7
"@" "A" "B" "C" "D" "E" "F" "G"
8 9 10 11 12 13 14 15
"H" "I" "J" "K" "L" "M" "N" "O"
16 17 18 19 20 21 22 23
"P" "Q" "R" "S" "T" "U" "V" "W"
24 25 26 27 28 29 30 31
"X" "Y" "Z" "[" "\\" "]" "^" "_"
32 33 34 35 36 37 38 39
"`" "a" "b" "c" "d" "e" "f" "g"
40 41 42 43 44 45 46 47
"h" "i" "j" "k" "l" "m" "n" "o"
48 49 50 51 52 53 54 55
"p" "q" "r" "s" "t" "u" "v" "w"
56 57 58 59 60 61 62 63
"x" "y" "z" "{" "|" "}" "~" "\177"
64 65
"\u0080" "\u0081" And then use this dictionary to encode the GAPDH counts:
[1] "D1E1A1B1A1B1C1Z1B1M1A1E1B1C1X1D1C2I2"Because there are more (and shorter) runs in the GAPDH count vector than for MYC, the degree of compression we achieve is smaller, from 17 KiB for the original count vector to 7.8 KiB for the character representation.
In this post, the example dictionaries encode counts up to 256 - but many single-cell experiments will contain larger integer counts than 256.
For those, we either need to create a larger dictionary, or we could opt to truncate larger values (e.g. represent counts >= 256 with the same character, losing information in the process).
When visualizing gene expression scores, we often log2 transform the raw or normalized counts, generating floating point numbers. If it is acceptable to lose precision, we can round these log2 transformed scores to the nearest integer and apply run-length encoding to them.
Because 2 to the power of 256 is a very large number (~ 1.16e+77), we would be able to represent even the highest observed scores But on the flip side, rounding the log2 transformed scores would obscure some differences between expression levels.
At this point, we are representing the original count vectors as character strings. Some of these string representation contain repeated patterns, which we can exploit to compress them even further.
The Lempel–Ziv–Welch algorithm was the first widely used universal data compression method and is e.g. used in the GIF image format.
At a high level, the algorithm identifies the longest repeated sequences (“phrases”) in a character string and maps them to shorter expressions in a dictionary.
There are great tutorials about the LZW algorithm and its implementation available on the web, e.g. this one. Here, I will create a basic implementation in R 3.
compress_lzw <- function(input_string) {
1 dictionary <- setNames(intToUtf8(1:256, multiple = TRUE),
intToUtf8(1:256, multiple = TRUE)
)
next_code <- 257
2 previous_pattern <- character(0)
3 output <- character(0)
4 for (char in strsplit(input_string, "")[[1]]) {
5 new_pattern <- paste0(previous_pattern, char)
6 if (new_pattern %in% names(dictionary)) {
previous_pattern <- new_pattern
7 } else {
output <- paste0(output, dictionary[[previous_pattern]])
dictionary[[new_pattern]] <- as.character(intToUtf8(next_code))
next_code <- next_code + 1
previous_pattern <- char
}
}
if (nzchar(previous_pattern)) {
output <- paste0(output, dictionary[[previous_pattern]])
}
return(output)
}previous_pattern variable. It is initialized as an empty string.
output variable will collect the compressed output and is eventually returned. It is also initialized as an empty string.
previous_pattern is still empty. But in subsequent loops, we extend the previous one with the new character to identify longer patterns.
previous_pattern with the current one.
previous_pattern and add it to the outputLet’s apply our LZW compression function to the run-length encoded MYC expression we generated above:
[1] "a3b1d1āăa4b3ć2a1ĊĆ1ĂĒ7ăcĒĈ6Ĉ8Ĉ9Ĉ1ĞėĆ2bĎđa5ĈĪħīĈĖĆ6ąa2ij5ĘďĈĥıĦĉĔĴ2ĘĠŁĤĚįĠ0ĮĆ3ĸ1ĸ6ōŃĸĪĒġĒōĿŔĹĒěĆğĎ9ōłĎŚ1ŖĉĺĠĺfĆĨĨŧēĽĝŋœĽĿ20eŭōĻćŴŅĕżŪĒĨ7ąĚijŰğŋĈ4őĆŚƋāĶĽŰĽƎĆİŨŋŎŕũŭƟĩłŗļıĸƘĩĽŽĖĎŐƥď0Əďŧŝžďĸ3ƇƠĤƵ5ijěČ8ƸŇĒʼnƐŊďƒƒ1ŚIJƀưƈƌſĺƵ7ƣĴƍƃƧƃď4ąĸŽǂǏŧǞƞƌ4aNjhƌĎǢaƵƴĘijĨ8ƺƩħǠijNjĿŧĻcĎƝǘĤƲ1ŽƵ2ŚųśȂďǺĚǠǙDžĈŷĜǜŦȕęĒķƐţĴŚƆƐźĴȝŚůƵȇăgǽŪĭƙũƭĠŧĝĎƚƬƛƓȒĆȳȞŃņȜĕňĠœijǖƻƩȮȿƉȾĴdžȌťɍěȀijƹȚīǗNjŒļȭȸƶƚȁŌǤĸƚůůȁɣƸdzȱīȢƮĴɅďťĨĬɰĽƾȫɀƻɊƯĂȀƗĞǶĿǼƣǠǠƘȽɛȊɷďȻɯɔƪƂĤĨ3ƅģȷŰǗǕŏǛȹȋȊ3ȪǃșaʞĢ1ȢɠƶűǒƶƒĥĎǼįȐĤǶɣơɤȘʣȱōɍțāŢŧɯŽǤȑȌŽʔȁȁ˂ĺijĵɆɞǡăŬʾ1ȪȋȅżʛƳȖĠȉƉġĎŽʭȖŧɏʕŽŷ2źơ1ƱǏńžʧʸˬȓȖĨ23˪ſȷƫœƏǗ˟ĉōˈȊ˧ʲƶ˷ōƵʨĔ̉˄ŖʚǣɍŽ̓ƕ˶Ȗŏɪęʯĺɍ3ǎƷǏȊƅʡư˒ĄƄŶ˫Njɑď˱ŅȰ˺ȔȿȌǀƶ̬˂ʿďɭŚƑơNjŧʨ̬ġʨŚɼaİĸƒʤʢ̤͋ǍƧǶȊ5ǫōźƇȀʨʭƉɯ˃LJĒʨˡȕČĿɭnj˄̄ǶʊǵɆ˳ŻŜɋȗŞȢĊɛŷɍˤăƔƠČƵƎɽ̳̱ͦơ̸ĤȢ̔ŭ˂5ͳȖͤ˫ɐʥɇɜȊş˖Ǽ̀Ίͯŋǡͺʩ̲ǩįƿȽĒĥČɷˌǜʉ\u03a2ɜƨ˸ȃƤȣʫ\u0378ŋƉ0ȎɕƯ6"After LZW compression, R estimates the size of this new character representation to be 1.4 KiB, down from 2.6 KiB after RLE.
Next, let’s implement the corresponding decoding function!
decompress_lzw <- function(compressed) {
1 dictionary <- setNames(intToUtf8(1:256, multiple = TRUE),
intToUtf8(1:256, multiple = TRUE)
)
next_code <- 257
2 compressed_codes <- strsplit(compressed, "")[[1]]
# read the first character and decode it
3 current_code <- compressed_codes[1]
output <- dictionary[[current_code]]
previous_string <- output
# read the remaining characters, one by one
4 for (code in compressed_codes[-1]) {
if (as.character(code) %in% names(dictionary)) {
5 current_string <- dictionary[as.character(code)]
dictionary[[intToUtf8(next_code)]] <- paste0(previous_string,
substr(current_string, 1, 1))
} else {
current_string <- paste0(previous_string, substr(previous_string, 1, 1))
dictionary[[intToUtf8(next_code)]] <- current_string
}
output <- paste0(output, current_string)
next_code <- next_code + 1
previous_string <- current_string
}
return(output)
}Let’s test the function by restoring the RLE-encoded MYC counts we compressed above:
[1] "a3b1d1a3b1a4b3a3b2a14b1a13b1a17b1c1a1b1a6b1a8b1a9b1a1b1a9b1c1a2b2a4b1a5b1a5b2a4b1a5b1a7b1a6d1a2d1a5c1a1b1a2b1a6b2a43b1a22c1b1a12c1a2b1a6b1a7b1a10b1a5b1a3c1a1c1a6c1a1b1a12c1a5b1a11b1a1c1a13b1a5b1a1b1a16b1a9b2a9c1a1c1b2a5b1a11b1a4b1a2b1a1b1a2f1a4b1a4b1a1b1a13b2a8b1a3c1a5b2a3b1a20e1a4c1a12b1a3b1a3c1a2b1a17c1a12b1a1b1a14b1a7d1b1a6d1a1b1a19b1a3b1a4c1a1b1a5b1a9b1a3d1a5b2a1b1a1b2a4c1a7b1a4b1a31c1a11b1a2b1a4b1a2b1a5c1b1a1c1a6b1a6c1a4c1a5b2a2b1a7b2a6c1a6b1a10c1a1b1a11b1a6b1a3b1a1c1a3d1b1a4b1a26b1a5d1a6b3a8c1a3b1a7b1a10b1a5b1a5b1a19b1a9b1a15b1a6d1a2b1a10b1a6d1a3b1a3c1b1a26b1a7c1b1a2b1a4b1a1b1c1a4b1a1b1a14d1c1a2b1a8c1a2b1a1b1a4d1a11b1a3b4a9b1a1h1a3b2a8c1a6b1a1b1a6c1d1a4b1a8d1b1a5b2a4c1a2d1a9b1a13b1a1b1a2b1c2a1c1a2b1a2c1a1b1a12b1a6b1a25b1a8b1a1b1a2b1a1d1a9b1a6c1a2b1a4b1a10b1a3b1a2b1a8b1a1b1a11b1a8b1a1b1a15c1a5c1b2a25b1a7d1a5e1a2c1b2a5b1a4b1a16b1a6b1a2b1g1b1a2b1a1b2a4b1a7b1a2b2a6b1a11b1a8b2a7b1a7b2a4b1a3d1a10b1a8b2a25b1a12b1a6b1a5c1a17b1a10b1a1c1a5d1a7c1a4b1a5b1a7b1a5c1d1a1b1a6b1a20b1a2b1a5b1a10b1a6b2a1d1a3d1a15b1a5c1b1a9b1a1b1a12c1a6b2a4b1a3d1a3b1a7b1a1c1a3c1b1a4c1a7b1a4b1a14b1a11c1a7b1a4c1a3d1a4b1a11b1a5e1a6c1a27c1a15b1a14b1a5b2a15b2a5d1b1a2b1a17b1a4b1d1a1b1a6b1a3b2a1b2a4b1a9d1b1a3b1a3b1a1c1b1c1a2c1a24c1b1a12b2a4b1a8b1a1b1a2b1a18b2a7c1a3d1b2a2c1a12b1a24b1a34b1a7b1c1a4b1a1b1a1c1b1a6b1a7c1a6c1a4b1a10b1a1b1a8b1a3g1c1a3b1a1b1a8b1a3b1a9b1e1a3c1a3b1a13b1a3b1a3b1a9b1a2b2a3b1a1b1a7c1a2b1a2d1b1a7b1a4b1a2b1a4b1a14b1a8b1a8b1a11c1a10b1a5c1a3c1a1c1b1a7c1a2b1a1b1a4b1a4b1a2b1a2b1a4b1a7b1c1a1c1a2b1a1b1a2d1a2d1a4b1a7b1a12b1a8b1f1a3c1g1a1b1a12c1a12c1a4b1a11b1a1b1a1b1a15b1a8d1a11b2a2b1a9b1a2b1a1b1a11b1a5b1a10b1c1a2b1a3b1a22e1b1a2b1a10c1a2b1a2c1a3b1e1a3b1a8b1a10b1a3b1a1b1a14b1a23e1b1a3c1a4b1a2b1a7c1a5c1a1b1c1b1a1b2a4c1a11c1a18b1a2b1a3c1a2b1a3b1a23c1a16b1a3c1a3b1c1a16b1a4b1a1b1c1a6c1a2b1a10b1a2b1a2b1a2b2a14b1a2b1a1b1a1c1a6b1a11b1a1b2a3b1a20b1a36d1a1c1a2b1a8b1a4b1a7c1a3b1a10f1a1d1a14b2a3b1a2b1a19b1a16b2a1e1a3b1a2b2a6b1a3c1a3b1a2b1a5c1a2b1a6b3a3b1a6b2a2b1a1c1a1c1a16c1a5b1a5b1a9b1a2b1a9b1a11b1a3c1a6b2a1b3c1a5b1a3b2a7b1c1a9b1a8b1a3b1a1b1a8b1a3b1c1a3b1a5b1a6c1a4d1b1a8b1a5h1c1a1e1d1b2a13c1a9b1a2d1a17c1a1b1a4b1a5b1a13c1a2b1a9b1a8b3a3b1a6c1a15b1a4b1a8b1a2d1b1a1b1a2b1a8d1a4b1a10b1a4c1a16b1a6b1a11b1a9e1a4b2a4b1a3b1a20b1a1b1a5b1d1a5b1a4b3a6b1a4c2a1b1a5c1a15b1a3c1a3b1a2b1a2b1a1c1a2e1a2b1a2b1a42b1a15e1a4b1a1b1a13b1a6b1a2b1a10b1a6b1a9b1a5b1a3d1a8b1a9b2a1b1a13b1a11b2a5e1a16b1a32b1a8d1a5b1a3b1a13b1a2b1a9b1a7d1a6b1a12b1a12b3a1b1a2b1d1a2b1a1b1a8b1a1b1a12b1a3d1a4c1a3e1a2c1a1c1a2c1a3b1a1b1a5b1a3d1a10d1a9b1a15b1a6b1a6"Are the results of sequention compression & decompression really identical to the input string?
Because it’s hard to visually compare the long vectors we have compressed & decompressed above, here is a toy example:
Original String: MARYHADALITTLELAMBMARYHADALITTLELAMBMARYHAD Compressed: MARYHADALITTLELAMBāăąćĉċčďđēĄĆ Decompressed: MARYHADALITTLELAMBMARYHADALITTLELAMBMARYHAD We have applied two rounds of compression to our count (or rounded, log2 transformed counts) single-cell RNA-seq data, achieving considerable reduction in the size of the objects 4.
While I have created examples of RLE and LZW compression using R, the same approach can be implemented in any programming language. The reports generated by Parse Bioscience, for example this one, include rounded log2 gene expression counts for all assayed genes across all cells that was compressed in the same way 5. To decode the compressed strings, the html reports include javascript functions that perform the same operations established here.
Combined with interactive plots, users can enter a gene name, triggering the decoding of its gene expression scores and updating a UMP plot in their browser - without the need for any central server. (Take a look at the source code of one of their reports to see how this is achieved!)
Now that I have a better understanding of how large amounts of data can be included in a stand-along HTML document, I am ready to learn more about javascript & interactive visualizations 6!
R version 4.4.1 (2024-06-14)
Platform: x86_64-pc-linux-gnu
Running under: Debian GNU/Linux 12 (bookworm)
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.21.so; LAPACK version 3.11.0
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
time zone: America/Los_Angeles
tzcode source: system (glibc)
attached base packages:
[1] stats4 stats graphics grDevices datasets utils methods
[8] base
other attached packages:
[1] TENxPBMCData_1.24.0 HDF5Array_1.34.0
[3] rhdf5_2.50.1 DelayedArray_0.32.0
[5] SparseArray_1.6.0 S4Arrays_1.6.0
[7] abind_1.4-8 Matrix_1.7-0
[9] SingleCellExperiment_1.28.1 SummarizedExperiment_1.36.0
[11] Biobase_2.66.0 GenomicRanges_1.58.0
[13] GenomeInfoDb_1.42.1 IRanges_2.40.1
[15] S4Vectors_0.44.0 BiocGenerics_0.52.0
[17] MatrixGenerics_1.18.0 matrixStats_1.4.1
loaded via a namespace (and not attached):
[1] KEGGREST_1.46.0 xfun_0.49 lattice_0.22-6
[4] rhdf5filters_1.18.0 vctrs_0.6.5 tools_4.4.1
[7] generics_0.1.3 curl_6.0.1 AnnotationDbi_1.68.0
[10] tibble_3.2.1 RSQLite_2.3.9 blob_1.2.4
[13] pkgconfig_2.0.3 dbplyr_2.5.0 lifecycle_1.0.4
[16] GenomeInfoDbData_1.2.13 compiler_4.4.1 Biostrings_2.74.1
[19] htmltools_0.5.8.1 yaml_2.3.10 pillar_1.10.0
[22] crayon_1.5.3 cachem_1.1.0 mime_0.12
[25] ExperimentHub_2.14.0 AnnotationHub_3.14.0 tidyselect_1.2.1
[28] digest_0.6.37 purrr_1.0.2 dplyr_1.1.4
[31] BiocVersion_3.20.0 fastmap_1.2.0 grid_4.4.1
[34] cli_3.6.3 magrittr_2.0.3 withr_3.0.2
[37] rappdirs_0.3.3 filelock_1.0.3 UCSC.utils_1.2.0
[40] bit64_4.5.2 rmarkdown_2.29 XVector_0.46.0
[43] httr_1.4.7 bit_4.5.0.1 png_0.1-8
[46] memoise_2.0.1 evaluate_1.0.1 knitr_1.49
[49] BiocFileCache_2.14.0 rlang_1.1.4 glue_1.8.0
[52] DBI_1.2.3 BiocManager_1.30.25 renv_1.0.11
[55] jsonlite_1.8.9 R6_2.5.1 Rhdf5lib_1.28.0
[58] zlibbioc_1.52.0 
This work is licensed under a Creative Commons Attribution 4.0 International License.
Both of these approaches are used e.g. in the summary HTML reports generated by Parse Bioscience’s analysis pipeline, e.g. in this example report. The HTML file contains transcriptome-wide gene expression scores for > 50,000 cells, and users can visualize the expression of their favorite gene on a UMAP projection. The whole HTML file is ~ 60 MB in size.↩︎
Because runs can be very long, the lengths - represented as integers - have varying number of decimals.↩︎
While this implementation works, it could be improved. For example, it doesn’t set any limits on the size of the final dictionary.↩︎
Typical file formats used to exchange single-cell RNA-seq count tables are using more sophisticated ways of compression. For example, applying gzip compression to CSV files with sparse count data will reduce their sizes tremendously. Other formats, e.g. HDF5 or Parquet files also take advantage of compression algorithms.↩︎
The encoding & compression is happening when the report is generated, but the decoding javascript functions are part of the HTML report.↩︎
Greg Wilson’s Javascript for Data Scientists course has been on my to-do list for a while.↩︎
---
title: "Compressing single-cell RNA-seq data with run-length encoding and LZW"
author: "Thomas Sandmann"
date: "2024-12-23"
execute:
echo: true
warning: false
message: false
freeze: true
categories: [R, single-cell, RNA-seq, algorithms, TIL]
editor:
markdown:
wrap: 72
format:
html:
anchor-sections: true
toc: true
toc-depth: 4
code-tools:
source: true
toggle: false
caption: none
editor_options:
chunk_output_type: console
---
```{r}
#| include: false
renv::use(lockfile = file.path(path.expand("~"), "repositories", "blog",
"posts", "lzw", "renv", "profiles", "lzw",
"renv.lock"))
```
## tl;dr
- Today I learned how to compress integer vectors using
[run-length encoding (RLE)](https://en.wikipedia.org/wiki/Run-length_encoding),
output the runs as character vectors, and create a basic implementation
of the
[Lempel-Ziv-Welch (LZW)](https://en.wikipedia.org/wiki/Lempel%E2%80%93Ziv%E2%80%93Welch)
compression algorithm.
- Together, these approaches reduce the amount of information that needs to be
stored, e.g. in HTML reports, for interactive visualizations.
- Hat tip to analysis reports generated by
[Parse Bioscience's analysis pipeline](https://support.parsebiosciences.com/hc/en-us/articles/27066395947412-How-Do-I-Analyze-my-Parse-Biosciences-Data) where I spotted compressed single-cell RNA-seq data for the
first time.
## Motivation
I am always looking for ways to communicate genomics analyses to collaborators
in a way that allows them to drill down into the data themselves. Stand-alone
HTML reports can be opened in any web browser and can support rich
visualizations, making them my an indespensible tool in my tool kit.
Yet, some of the data I am working with, e.g. single-cell RNA-seq data,
is pretty large, and the size of my HTML reports - which include the data
as text - can quickly balloon. Luckily, there are ways to compress numerical
data without loosing information.
Here, I am exploring two approaches:
[run-length encoding (RLE)](https://en.wikipedia.org/wiki/Run-length_encoding) and
[Lempel-Ziv-Welch (LZW) compression](https://en.wikipedia.org/wiki/Lempel%E2%80%93Ziv%E2%80%93Welch)[^1].
[^1]: Both of these approaches are used e.g. in the summary HTML reports
generated by Parse Bioscience's analysis pipeline, e.g. in this
[example report](https://cdn.parsebiosciences.com/mouse/brain-nuclei/v3/all-sample_analysis_summary.html#).
The HTML file contains transcriptome-wide gene expression scores for > 50,000
cells, and users can visualize the expression of their favorite gene on a
UMAP projection. The whole HTML file is ~ 60 MB in size.
## Sparse information: single-cell RNA-seq data
Single-cell RNA-seq data are notoriously sparse, e.g. the majority of counts
across genes & cells is zero. For example, 96% of all counts in the PBMC 4k
single-cell experiment shared by 10X Genomics (and available via the
[TENxPBMCData](https://bioconductor.org/packages/release/data/experiment/html/TENxPBMCData.html)
Bioconductor package) are zero:
```{r}
#| message: false
#| warning: false
library(TENxPBMCData)
tenx_pbmc4k <- suppressMessages(TENxPBMCData(dataset = "pbmc4k"))
mean(counts(tenx_pbmc4k) == 0)
```
Most of the remaining counts are integers smaller than ten, with fewer than
0.2% of values exceeding that range:
```{r}
mean(counts(tenx_pbmc4k) > 10)
```
This leads to very long runs of identical integers (most often zero), which
can efficiently be represented through
[run-length encoding](https://en.wikipedia.org/wiki/Run-length_encoding)
For example, the expression of the transcription facto _MYC_ gene is highly
skewed towards zero counts:
```{r}
myc <- local({
ensembl <- row.names(tenx_pbmc4k)[which(rowData(tenx_pbmc4k)$Symbol == "MYC")]
counts(tenx_pbmc4k)[ensembl, ]
})
hist(myc, breaks = 100, xlab = "MYC (raw counts)")
```
and there are long sequences of zeros and some shorter sequences of ones in
the count vector. (Here, I am only showing counts for the the first 100 of the
`r ncol(tenx_pbmc4k)` cells.)
```{r}
head(myc, n = 100)
```
R estimates that the entire vector of `r length(myc)` counts occupies
`r format(object.size(myc), units = "KiB")` of memory.
## Run length encoding
This information can be communicated more concisely by noting each integer (e.g.
zero) and the number of times it is repeated. The `stats::rle` function
performs this conversion:
```{r}
run_lengths <- rle(myc)
run_lengths
```
and returns two components:
1. a vector of values:
```{r}
head(run_lengths$values)
```
2. a vector with the number of times each of them is repeated:
```{r}
head(run_lengths$lengths)
```
This allows us to compress the original `r ncol(tenx_pbmc4k)`
integers into `r sum(lengths(run_lengths))` integers
(`r lengths(run_lengths)[1]` length & value pairs),
without losing any information.
By combining information about values and run-lengths, we can reconstruct
the original vector, either manually:
```
0 x 3: 0,0,0,
1 x 1: 1,
3 x 1: 3,
0 x 3: 0,0,0,
1 x 1: 1,
0 x 4: 0,0,0,0,
1 x 3: 1,1,1,
...
=> 0,0,0,1,3,0,0,0,1,0,0,0,0,1,1,1,...
```
or via `inverse.rle`:
```{r}
head(inverse.rle(run_lengths), n = 16)
```
To store run-length encoded values e.g. in a text (or HTML) file, we can also
represent them as characters, similar to our manual reconstruction above, e.g.
separating values and lengths with an `x`:
```{r}
myc_rle_separated <- mapply(paste,
run_lengths$values,
run_lengths$length,
MoreArgs = list(sep = "x")) |>
paste(collapse = ";")
substr(myc_rle_separated, 1, 40)
```
This representation occupies only
`r format(object.size(myc_rle_separated), units = "KiB")` of memory - but _half_
of the character string is required to encode the `x` and `;` separators. We
can do better!
To avoid including any separators, we can represent one of the
numbers, e.g. the values, as a single character instead. For the _MYC_ gene, the
largest observed integer count is `r max(myc)`. So let's represent all
`r max(myc) + 1` values (including 0) with a letter of the alphabet instead:
```{r}
dictionary <- setNames(letters[1:(max(myc) + 1)], 0:max(myc))
dictionary
```
For example, the letter `a` represents `0`, `b` represents `1` and so on.
Now we can omit the `x` and `;` separators, as each run is unambiguously
represented by one character and an integer [^2]:
```{r}
myc_rle_character <- mapply(paste0,
dictionary[as.character(run_lengths$values)], # <1>
run_lengths$length) |>
paste(collapse = "")
substr(myc_rle_character, 1, 21)
```
1. It's critical to coerce the run-length values into a _character_ vector here,
otherwise the named vector is subset by _position_ instead of by _name_.
[^2]: Because runs can be very long, the lengths - represented as integers -
have varying number of decimals.
In this representation, the `r ncol(tenx_pbmc4k)` counts value for the _MYC_
gene now occupy only `r format(object.size(myc_rle_character), units = "KiB")`
of memory instead of the original `r format(object.size(myc), units = "KiB")`.
We can also write the data to a text (or HTML) file in this format, e.g. as a
single string.
::: {.callout-note}
### Encoding larger numbers of values
In this example, we only needed `r max(myc) + 1` letters to encode all observed
unique counts for the _MYC_ gene. But for other genes, especially those
expressed at higher levels, we might observe many more possible values - even
more than the `r length(letters)` lower case letters of the English alphabet.
```{r}
gapdh <- local({
ensembl <- row.names(tenx_pbmc4k)[
which(rowData(tenx_pbmc4k)$Symbol == "GAPDH")]
counts(tenx_pbmc4k)[ensembl, ]
})
```
For example, in this dataset we observe `r length(table(gapdh))` distinct
count values for the highly abundant _GAPDH_ gene:
```{r}
table(gapdh)
```
Luckily, we are not restricted to the English alphabet. For example, we can map
to any character in the
[Unicode Transformation Format – 8-bit (UTF-8)](https://en.wikipedia.org/wiki/UTF-8)
character encoding instead.
To encode e.g. all integers from 1 to 256 (more than enough for _GAPDH_) - but
*avoiding those that represent a space, quote, etc or integers* -
we can use UTF-8 codes 64 to 319:
```{r}
codes <- 64:319
dictionary <- setNames(intToUtf8(codes, multiple = TRUE), 0:255)
head(dictionary, n = 66)
```
And then use this dictionary to encode the _GAPDH_ counts:
```{r}
head(gapdh, n = 20)
```
```{R}
gapdh_character <- local({
run_lengths <- rle(gapdh)
mapply(paste0,
dictionary[as.character(run_lengths$values)],
run_lengths$length) |>
paste0(collapse = "")
})
substr(gapdh_character, 1, 36)
```
Because there are more (and shorter) runs in the _GAPDH_ count vector than
for _MYC_, the degree of compression we achieve is smaller, from
`r format(object.size(gapdh), units = "KiB")` for the original count vector to
`r format(object.size(gapdh_character), units = "KiB")` for the character
representation.
:::
::: {.callout-caution collapse="true"}
In this post, the example dictionaries encode counts up to 256 - but many
single-cell experiments will contain larger integer counts than 256.
For those, we either need to create a larger dictionary, or we could opt to
truncate larger values (e.g. represent counts >= 256 with the same character,
losing information in the process).
When visualizing gene expression scores, we often log2 transform the raw or
normalized counts, generating floating point numbers. If it is acceptable to
lose precision, we can round these log2 transformed scores to the nearest
integer and apply run-length encoding to them.
Because 2 to the power of 256 is a very large number (~ 1.16e+77), we would
be able to represent even the highest observed scores But on the flip side,
rounding the log2 transformed scores would obscure some differences between
expression levels.
:::
## LZW compression
At this point, we are representing the original count vectors as character
strings. Some of these string representation contain repeated patterns, which
we can exploit to compress them even further.
::: {.callout-note collapse="true"}
The `Lempel–Ziv–Welch` algorithm was the
[first widely used universal data compression method](https://en.wikipedia.org/wiki/Lempel%E2%80%93Ziv%E2%80%93Welch)
and is e.g. used in the GIF image format.
At a high level, the algorithm identifies the longest repeated sequences
("phrases") in a character string and maps them to shorter expressions in a
dictionary.
:::
There are great tutorials about the LZW algorithm and its implementation
available on the web, e.g.
[this one](https://eng.libretexts.org/Bookshelves/Electrical_Engineering/Signal_Processing_and_Modeling/Information_and_Entropy_(Penfield)/03%3A_Compression/3.07%3A_Detail-_LZW_Compression/3.7.01%3A_LZW_Algorithm_Example_1).
Here, I will create a basic implementation in R [^3].
[^3]: While this implementation works, it could be improved. For example, it
doesn't set any limits on the size of the final dictionary.
### LZW encoding
```{r}
compress_lzw <- function(input_string) {
dictionary <- setNames(intToUtf8(1:256, multiple = TRUE), # <1>
intToUtf8(1:256, multiple = TRUE)
)
next_code <- 257
previous_pattern <- character(0) # <2>
output <- character(0) # <3>
for (char in strsplit(input_string, "")[[1]]) { # <4>
new_pattern <- paste0(previous_pattern, char) # <5>
if (new_pattern %in% names(dictionary)) { # <6>
previous_pattern <- new_pattern
} else { # <7>
output <- paste0(output, dictionary[[previous_pattern]]) # <7>
dictionary[[new_pattern]] <- as.character(intToUtf8(next_code))
next_code <- next_code + 1
previous_pattern <- char
}
}
if (nzchar(previous_pattern)) {
output <- paste0(output, dictionary[[previous_pattern]]) # <8>
}
return(output)
}
```
1. We initialize a "dictionary" (e.g. a named character vector) with 256 UTF-8
codes.
2. As we parse the input string, the current sequence will be stored in the
`previous_pattern` variable. It is initialized as an empty string.
3. The `output` variable will collect the compressed output and is
eventually returned. It is also initialized as an empty string.
4. The input string is split into individual characters, and we process them
one by one.
5. In the first loop, the `previous_pattern` is still empty. But in subsequent
loops, we extend the previous one with the new character to identify longer
patterns.
6. If the extended pattern is already known, then we don't need to update our
dictionary. We simply replace the `previous_pattern` with the current one.
7. If we encounter a _new_ pattern, then we
- Encode the `previous_pattern` and add it to the output
- Map the new pattern to the next available UTF-8 code
- Increment the position of the next available UTF-8 code by one
- Reset the previous pattern to the current character.
8. After the entire string has been processed, we encode the final pattern
and then return the full output.
Let's apply our LZW compression function to the run-length encoded _MYC_
expression we generated above:
```{r}
#| message: false
myc_lzw <- compress_lzw(myc_rle_character)
myc_lzw
```
After LZW compression, R estimates the size of this new character representation
to be `r format(object.size(myc_lzw), units = "KiB")`, down from
`r format(object.size(myc_rle_character), units = "KiB")` after RLE.
Next, let's implement the corresponding decoding function!
### LZW decoding
```{r}
decompress_lzw <- function(compressed) {
dictionary <- setNames(intToUtf8(1:256, multiple = TRUE), # <1>
intToUtf8(1:256, multiple = TRUE)
)
next_code <- 257
compressed_codes <- strsplit(compressed, "")[[1]] # <2>
# read the first character and decode it
current_code <- compressed_codes[1] # <3>
output <- dictionary[[current_code]]
previous_string <- output
# read the remaining characters, one by one
for (code in compressed_codes[-1]) { # <4>
if (as.character(code) %in% names(dictionary)) {
current_string <- dictionary[as.character(code)] # <5>
dictionary[[intToUtf8(next_code)]] <- paste0(previous_string,
substr(current_string, 1, 1))
} else {
current_string <- paste0(previous_string, substr(previous_string, 1, 1))
dictionary[[intToUtf8(next_code)]] <- current_string
}
output <- paste0(output, current_string) # <6>
next_code <- next_code + 1 # <7>
previous_string <- current_string # <8>
}
return(output)
}
```
1. The decoding function is provided with the same initial dictionary we
included in the encoding function.
2. We process the compressed string one character at a time.
3. The algorithm is initialized with the first character of the encoded string,
and we decode it using the pre-defined dictionary (which includes all single
characters).
4. We then iterate over the remaining characters in the compressed input.
5. We check if the current code is already in our dictionary
- If _yes_, then we decode it by looking up its corresponding value in our
dictionary. We also add an extended pattern to our dictionary, by
concatenating the previous result with the (first character of) our
current one.
- If _no_, then we decode it by extending the previous result by its (own)
first character and add it to the dictionary.
character
6. We add the newly decoded result to our output string.
7. Increment the running index of our dictionary, because we added a new key.
8. Get ready for the next iteration.
Let's test the function by restoring the RLE-encoded _MYC_ counts we
compressed above:
```{r}
myc_lzw_decompressed <- decompress_lzw(myc_lzw)
head(myc_lzw_decompressed)
```
Are the results of sequention compression & decompression really identical
to the input string?
```{r}
identical(
decompress_lzw(compress_lzw(myc_rle_character)),
myc_rle_character)
```
::: {.callout-note collapse="true"}
#### Compression / decompression example
Because it's hard to visually compare the long vectors we have compressed &
decompressed above, here is a toy example:
```{r}
original_string <- "MARYHADALITTLELAMBMARYHADALITTLELAMBMARYHAD"
compressed <- compress_lzw(original_string)
decompressed <- decompress_lzw(compressed)
```
```{r}
#| echo: false
cat("Original String: ", original_string, "\n")
```
```{r}
#| echo: false
cat("Compressed: ", compressed, "\n")
```
```{r}
#| echo: false
cat("Decompressed: ", decompressed, "\n")
```
:::
## So what?
We have applied two rounds of compression to our count (or rounded, log2
transformed counts) single-cell RNA-seq data, achieving considerable reduction
in the size of the objects [^4].
[^4]: Typical file formats used to exchange single-cell RNA-seq count tables
are using more sophisticated ways of compression. For example, applying
[gzip compression](https://en.wikipedia.org/wiki/Gzip) to CSV files with
sparse count data will reduce their sizes tremendously. Other formats, e.g.
`HDF5` or `Parquet` files also take advantage of compression algorithms.
While I have created examples of RLE and LZW compression using R, the same
approach can be implemented in any programming language. The reports generated
by Parse Bioscience, for example
[this one](https://cdn.parsebiosciences.com/mouse/brain-nuclei/v3/all-sample_analysis_summary.html#),
include rounded log2 gene expression counts for all assayed genes across all
cells that was compressed in the same way [^5]. To decode the compressed
strings, the html reports include javascript functions that perform the same
operations established here.
[^5]: The encoding & compression is happening when the report is generated,
but the decoding javascript functions are part of the HTML report.
Combined with interactive plots, users can enter a gene name, triggering the
decoding of its gene expression scores and updating a UMP plot in their
browser - without the need for any central server. (Take a look at
[the source code of one of their reports](https://cdn.parsebiosciences.com/mouse/brain-nuclei/v3/all-sample_analysis_summary.html#)
to see how this is achieved!)
Now that I have a better understanding of how large amounts of data can be
included in a stand-along HTML document, I am ready to learn more about
javascript & interactive visualizations [^6]!
[^6]: Greg Wilson's
[Javascript for Data Scientists](https://third-bit.com/js4ds/)
course has been on my to-do list for a while.
## Reproducibility
<details>
<summary>
Session Information
</summary>
```{r}
sessionInfo()
```
</details>